Oral Paper Presentation
Annual Scientific Meeting
Session: Plenary Session 3A - Liver
40 - Gender Disparity in Hepatocellular Carcinoma Survival: Exploiting Tissue-Specific Protein Interaction Networks to Unveil Prognostic Gene Modules
Tuesday, October 24, 2023
2:45 PM – 2:55 PM PT
Location: Ballroom A

Basile Njei, MD, MPH, PhD
Yale School of Medicine
New Haven, CT
Presenting Author(s)
Lydia Fozo, BS1, Basile Njei, MD, MPH, PhD2
1Harvard University, Boston, MA; 2Yale School of Medicine, New Haven, CT
Introduction: Men are three times more likely to develop Hepatocellular Carcinoma (HCC) and often have poorer responses to treatment. Our study seeks to identify sex-specific disease gene modules that characterize the molecular mechanisms sponsoring differential survival outcomes in males vs. females.
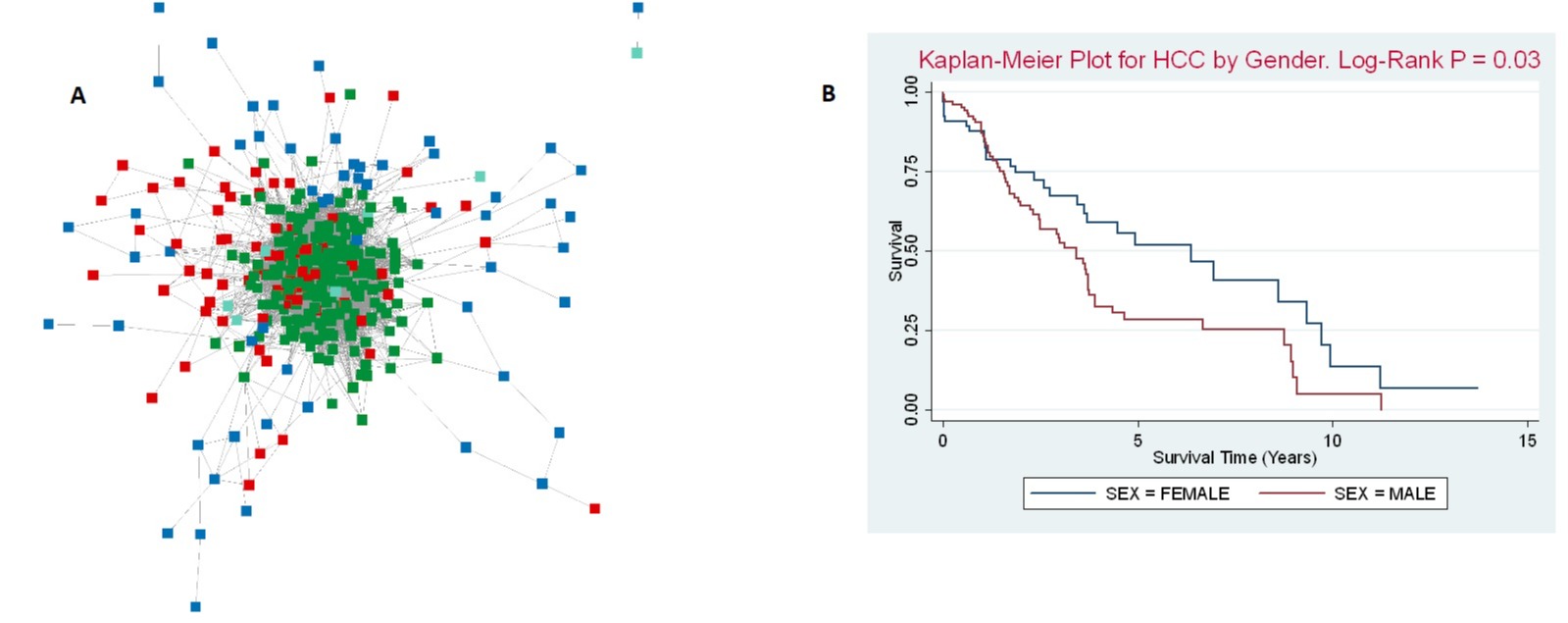
Methods: We employed a series of computational methods to investigate the molecular characteristics of HCC. First, we constructed a protein-protein interaction (PPI) network using validated protein interactions in liver tissue. Next, we employed a widely used algorithm called DIseAse MOdule Detection (DIAMOnD) to identify disease modules within the PPI network. To focus on HCC specific gene activity, we used a curated set of genes associated with cell state switching in HCC, as well as previously identified HCC disease genes, as seed genes in the PPI network. This integration allowed us to identify a novel set of 230 potential disease genes, forming a DIAMOnD gene module (Fig. 1A).These submodules represent groups of genes with high interconnectedness, suggesting important functional clusters within the larger module. We conducted Cox regression analysis using the The Cancer Genome Atlas Liver Hepatocellular Carcinoma gene expression dataset.
Results: We included 196 HCC patients in our analytic samples. Included patients were mostly male (66%), median age was 62 years, and 60% were non-Hispanic white. Male gender was associated with poorer survival (Fig. 1B). In a multivariable model adjusting for stage, DNA hypermethylation clusters, male gender was associated with a 60% increased risk of all-cause mortality (aHR=1.59; 95% CI: 1.02–2.48, p=0.04).
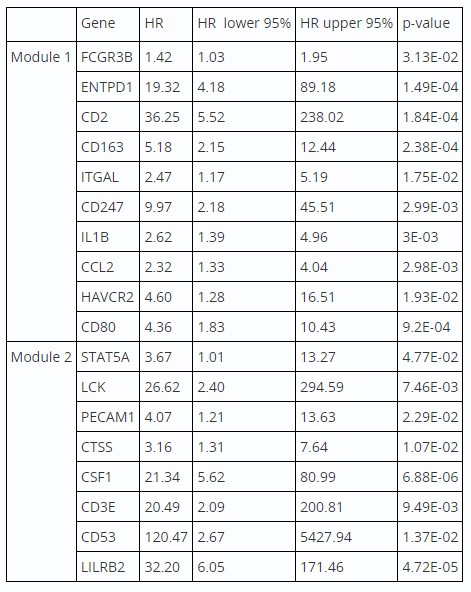
Furthermore, within the top two submodules, namely Module 1 and Module 2, we identified 10 and 8 genes, respectively, that demonstrated a significant association with adverse prognosis specifically in males (Table 1).
Discussion: Our findings reveal distinct gene modules that contribute to the differential survival observed between males and females. These findings highlight the importance of gender-specific considerations in HCC prognosis and provide insights into potential molecular mechanisms driving the observed disparities. Future studies should aim to explore the underlying molecular mechanisms driving these gene modules.
Disclosures:
Lydia Fozo, BS1, Basile Njei, MD, MPH, PhD2, 40, Gender Disparity in Hepatocellular Carcinoma Survival: Exploiting Tissue-Specific Protein Interaction Networks to Unveil Prognostic Gene Modules, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.
1Harvard University, Boston, MA; 2Yale School of Medicine, New Haven, CT
Introduction: Men are three times more likely to develop Hepatocellular Carcinoma (HCC) and often have poorer responses to treatment. Our study seeks to identify sex-specific disease gene modules that characterize the molecular mechanisms sponsoring differential survival outcomes in males vs. females.
Methods: We employed a series of computational methods to investigate the molecular characteristics of HCC. First, we constructed a protein-protein interaction (PPI) network using validated protein interactions in liver tissue. Next, we employed a widely used algorithm called DIseAse MOdule Detection (DIAMOnD) to identify disease modules within the PPI network. To focus on HCC specific gene activity, we used a curated set of genes associated with cell state switching in HCC, as well as previously identified HCC disease genes, as seed genes in the PPI network. This integration allowed us to identify a novel set of 230 potential disease genes, forming a DIAMOnD gene module (Fig. 1A).These submodules represent groups of genes with high interconnectedness, suggesting important functional clusters within the larger module. We conducted Cox regression analysis using the The Cancer Genome Atlas Liver Hepatocellular Carcinoma gene expression dataset.
Results: We included 196 HCC patients in our analytic samples. Included patients were mostly male (66%), median age was 62 years, and 60% were non-Hispanic white. Male gender was associated with poorer survival (Fig. 1B). In a multivariable model adjusting for stage, DNA hypermethylation clusters, male gender was associated with a 60% increased risk of all-cause mortality (aHR=1.59; 95% CI: 1.02–2.48, p=0.04).
Furthermore, within the top two submodules, namely Module 1 and Module 2, we identified 10 and 8 genes, respectively, that demonstrated a significant association with adverse prognosis specifically in males (Table 1).
Discussion: Our findings reveal distinct gene modules that contribute to the differential survival observed between males and females. These findings highlight the importance of gender-specific considerations in HCC prognosis and provide insights into potential molecular mechanisms driving the observed disparities. Future studies should aim to explore the underlying molecular mechanisms driving these gene modules.

Figure: Figure 1A. Cytoscape network of liver cancer switch genes (blue), known liver cancer disease genes (red), and disease genes extracted from DIAMOnD algorithm ( green).Network was generated using protein interactions from STRING-DB with turquoise nodes representing non DIAMoND genes with StringR interactions .
Figure 1B. Kaplan-Meier Survival analysis of HCC
Figure 1B. Kaplan-Meier Survival analysis of HCC

Table: Table 1: Cox regression summary results showing the hazard ratios and statistical significance of selected genes in relation to survival outcomes in Males.
Disclosures:
Lydia Fozo indicated no relevant financial relationships.
Basile Njei indicated no relevant financial relationships.
Lydia Fozo, BS1, Basile Njei, MD, MPH, PhD2, 40, Gender Disparity in Hepatocellular Carcinoma Survival: Exploiting Tissue-Specific Protein Interaction Networks to Unveil Prognostic Gene Modules, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.
